Ileana Martínez-Cabrera,1 Silvia Patricia Ortega-Moya,2, 3 Margalida Calafat-Sard,4 Carlos Dolz- Abadía,3, 4 Víctor José Asensio-Landa,3 Marco Bauzá-Thorbrügge,1, 3 Jordi Oliver-Oliver,1, 3, 5, 6 Carmen Hermida-Díaz,7 Francisco José García-Palmer1, 3, 5
1Grupo Metabolismo Energético y Nutrición, y IUNICS, Universitat de les Illes Balears.
2Hospital of Inca, Illes Balears.
3Instituto de Investigación Sanitaria de Baleares (IdISBa), Hospital Universitario Son Espases, edificio S. E-07120 Palma de Mallorca, Illes Balears.
4Hospital Son Llàtzer, Palma de Mallorca.
5Ciber Fisiopatología Obesidad y Nutrición (CB06/03) Instituto Salud Carlos III, Madrid.
6Grupo Multidisciplinar de Oncología Traslacional, y IUNICS, Universitat de les Illes Balears.
7Venter Pharma S.L., Alcobendas, Madrid, España.
Acta Gastroenterol Latinoam 2019;49(2):96-109
Recibido: 13/04/2018 / Aprobado: 13/08/2018 / Publicado en www.actagastro.org el 17/06/2019
Summary
Gut bacterial characterization could be a good criterion for prevention, diagnosis confirmation and monitoring of digestive diseases. Aim. To begin the first step for future establishment of the metagenomic methodology in Balearic healthy system for the characterization gut microbiota, in colorectal exploration of patients affected by bowel diseases. Material and methods. The bacteria were characterized in colorectal biopsy and brushing (healthy and inflamed regions) from the bowel by 16S rRNA gene V3-V4 regions and Nextera XT metagenomic techniques (Illumina). DNA was isolated by colonoscopy exploration from 10 subjects (20-75 years, both sex) who felt some symptoms of abdominal pain, diarrhea, episodic rectal bleeding. Gene libraries were prepared and sequenced for 451 bacterial genera. We evaluated the qualitative of food consumption as a complement. The results were analyzed by SPSS v.21. Results. Our results showed that the quality control of libraries and sequencing was adequate. There was great bacterial variability among subjects and sampling areas. PCA obtained showed that, the microbiota of patients was clustered together by disease type and the bacteria diversity of No evidence of active pathology patients clustered closely to ulcerative colitis distribution. Our preliminaries results suggested that the colorectal exploration is not enough for a complete diagnosis and follow-up of treatments. Conclusions. The first step to establish gut microbiota metagenomic technology in our group allowed us to obtain bacteria genomic libraries by colonoscopy exploration; from patient with bowel diseases; according to Illumina requirements. Preliminary results will permit to introduce the methodology; increasing the subject number and samples and nutritional characterization.
Key words. Gut microbiota, ulcerative colitis, Crohn´s disease, polyps, diverticula.
Microbiota intestinal: caracterización en casos de pacientes con trastornos digestivos activos
Resumen
La caracterización de la microbiota intestinal podría ser un buen criterio para la prevención, confirmación y seguimiento de las enfermedades digestivas. Objetivo. Comenzar la primera etapa para el establecimiento futuro de la metodología metagenómica para la caracterización de la microbiota intestinal en el sistema sanitario balear, por exploración colonoscópica en pacientes con enfermedades intestinales. Material y métodos. Las bacterias aisladas de biopsia y cepillado (regiones sana e inflamada) fueron cuantificadas utilizando las técnicas metagenómicas 16S rRNA V3-V4 y Nextera XT (Illumina), usando ADN bacteriano de diez sujetos (20-75 años, de ambos sexos) con síntomas de problemas digestivos (diarreas, dolor abdominal y sangrado rectal). Las librerías genómicas fueron preparadas y secuenciadas para 451 géneros de bacterias. Se evaluó el consumo cualitativo nutricional como información complementaria. Los resultados se analizaron utilizando SPSS v21. Resultados. Nuestros resultados mostraron un adecuado control de calidad de las librerías y su secuenciación. El análisis de componente principal mostró que la diversidad bacteriana se distribuyó por tipo de enfermedad y la distribución de los pacientes sin patología activa estuvo estrechamente relacionada con colitis ulcerosa. Estos resultados preliminares sugieren que la exploración colorrectal no es suficiente para el diagnóstico y seguimiento de los tratamientos. Conclusiones. La primera etapa para el establecimiento en nuestro grupo de la tecnología metagenómica de microbiota intestinal permitió obtener librerías genómicas a partir de exploración colonoscópica de pacientes con enfermedades intestinales; de acuerdo con los requerimientos de Illumina y permitirán introducir la metodología, incrementando el número de sujetos y muestras y su caracterización nutricional.
Palabras claves. Microbiota intestinal, colitis ulcerosa, enfermedad de Crohn, pólipos, divertículos.
Society advances, the responsibility, stress, changes in nutritional patterns when the people move for different geographical situations or adapt to fast food, stop consuming local products or simply, do not pay attention to their possible predisposition to digestive diseases; these and others constitute risk factors for inflammatory digestive diseases linked not only to the genetic-hereditary component, but also to the lifestyle and the socio-economic situation.
Gut microbiota is associated to a large number of inflammatory and metabolic diseases. It is evidenced by supporting the important functional role of gut in maintaining individual health, in the body as a whole.1
Most studies have suggested a great variability among individuals2 and among gastrointestinal tract areas in the same subject.3 Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria are the most abundant phyla in the human bowel4-6 Although gut microbiota pattern differs among subjects, there is a functional “core” of bacteria that it is necessary for a normal human host´s metabolism, some of them are represented in rural and urban population around the world; for example: Faecalibacterium, Roseburia, Bacteroides, Dorea, Clostridium, Eubacterium, Coprococcus, cepa de R., Alistipes, Collinsella, Parabacteroides and Bifidobacterium.7
Massive 16S rRNA gene sequencing technology (Illumina platform) has allowed bacterial identification in complex samples without the bacteria isolation.8
In Majorca, gut bacterial characterisation of colorectal biopsies in a healthy group (61-76 years) and Crohn´s disease patients (20-64 years) were performed and the microbiome cores were compared.9
Some analysis in ulcerative colitis (UC) patients demonstrated the difference between the luminal and mucus gel microbiota in control and in UC groups, with no differences among colorectal regions. No correlation was found between regional inflammation and a breakdown in this spatial or bacterial diversity.3
The aim of our work was to begin the first step for the future establishment of the methodology (with the collaboration of research Balearic units) for the massive metagenomic characterization of gut microbiota, in colorectal exploration of patients affected by bowel diseases.
Material and methods
Subjects and samples
Subjects without intestinal inflammatory background attended for abdominal pain, diarrhea, mucus secretion and episodic rectal bleeding, at the moment of exploration. They had not taken anti-inflammatory, immunosuppressive and antibiotic drugs in the previous 3 months and were explored by colonoscopy. Clinical indication, colonoscopy and histopathological findings were the inclusion criteria (Table 1). Individuals were included in five small groups: No evidence of active pathology (NEAP) (n = 2), a subject with diverticula (Diver), ulcerative colitis (UC, n = 2), Crohn´s disease (CD, n = 2) and polyps (polyps, n = 3) to demonstrate quality control of procedures: DNA extraction, libraries preparation and fidelity in sequencing process.
Samples were collected after an overnight fast with bowel preparations using polyethylene glycol, a day before the analysis. All patients received a dose of anesthesia. Mucosal biopsies and brushing samples were taken at 10 cm from affected areas and healthy opposite areas in patients; and from area in subjects without affected mucosa (apparently healthy) by colonoscopy exploration. One biopsy was placed into a sterile, nuclease-free tube (2 mL, Brand) with 1.5 mL of buffer InhibitEX (QIAamp Fast DNA Stool Mini Kit, QIAGEN) and one brush sample was placed in another sterile tube. Both were stirred and frozen at – 20 °C. A biopsy sample was fixed for histopathological assessment and analyzed according to protocols of both hospitals (Hospital of Inca and Hospital Son Llàtzer). Table 1 shows the clinical indications, colonoscopy and histopathology results as inclusion criteria for subject selection. NEAP subjects had no evidences of active pathology.
Table 1. Antecedents and exploration results (colonoscopy and histopathology analysis).
Samples:
Polyp group: -Pol1 [sample 1, healthy area brush (HABr); [sample 2, healthy area biopsy (HABio)]; [sample 3, inflamed area brush (IABr)]; [sample 4, inflamed area brush (IABio)]. -Pol2 [sample 5, healthy area brush (HABr); [sample 6, healthy area biopsy (HABio)]; [sample 7, inflamed area brush (IABr)]; [sample 8, inflamed area brush (IABio)]. -Pol3 [sample 15, healthy area brush (HABr); [sample 16, healthy area biopsy (HABio)]; [sample 17, inflamed area brush (IABr)]; [sample 18, inflamed area brush (IABio)].
UC: -UC1 [sample 11, healthy area brush (HABr); [sample 12, healthy area biopsy (HABio)]; [sample 13, inflamed area brush (IABr)]; [sample 14, inflamed area brush (IABio)]. -UC2 [sample 21, inflamed area brush (IABr)]; [sample 22, inflamed area brush (IABio)].
CD: -CD1 [sample 25, healthy area brush (HABr); [sample 26, healthy area biopsy (HABio)]; [sample 27, inflamed area brush (IABr)]; [sample 28, inflamed area brush (IABio)]. -CD2 [sample 29, healthy area brush (HABr); [sample 30, healthy area biopsy (HABio)]; [sample 31, inflamed area brush (IABr)]; [sample 32, inflamed area biopsy (IABio)].
NEAP: -H1 [sample 9, healthy area brush (HABr); [sample 10, healthy area biopsy (HABio)].-H2 [sample 23, healthy area brush (HABr); [sample 24, healthy area biopsy (HABio)].
Diverticula: -Diver1 [sample 19, healthy area brush (HABr); [sample 20, healthy area biopsy (HABio)].
Total Bacterial DNA extraction from samples
Bacterial DNA was extracted using the QIAamp® DNA Stool for Pathogen Detection Method Mini Kit (QIAGEN, Spain). The procedure was similar to the one described by manufacturer Manual User, adjusted to the samples in 1.5 mL buffer InhibitEX. The volume was processed completely.
DNA samples were concentrated by Concentrator plus equipment (Eppendorf), at 45 °C, for 95 min, until reaching a final volume of 40 μL. The amount of DNA, previously diluted with 10 mM Tris-HCl pH 8.5, was evaluated by protocol of QuantiFluor dsDNA KIT (Promega).
Analysis of bacterial content were performed by Genomic Libraries techniques (Illumina platform).
Genomic libraries were performed and sequenced (variable V3 and V4 regions of the 16S rRNA gene), using Amplicon 16S and Nextera XT Metagenomic (tagmentation reaction) methodologies (Illumina). We included a positive control of bacterial DNA and water free of RNAse/DNAse as a negative control.
An account of the results was created in BaseSpace platform and a series of results analysis applications were accessed on the Illumina website: http://www.illumina. com/index-d.html. 16S Metagenomics application performed the taxonomic classification of generated 16S rRNA amplification readings, using an Illumina cured version of the Greengenes taxonomic database. The classification was performed using the Illumina 16S Metagenomic workflow, which was also available in the MiSeq Reporter software. We evaluated the quality control of libraries, according to the references of Illumina; sensibility by massive sequencing and precision between Amplicon 16S and Nextera XT Metagenomic.
Food consumption pattern
We elaborated a questionnary according to the indications from Spanish Healthy Eating Guide,10 Norte and Ortiz;11 using the criteria about the frequency of consumption for ten general groups (grain and derivate, vegetables, fruit, dairy products, meat, cold-processed meat, legumes, sweets, beverages, and diet variety);11 by four days (including weekend), in the month before. Healthy Eating Index (HEI or IASE for Spain) was measured. The IASE categories (maximum score: 100) were: healthy feeding > 80, need-for-changes: 50-80, little healthy: < 50.11
Statistics analysis of results
Principal components analysis (PCA) and OTU (operational taxonomy unit) were applied to the DNA copies for 451 genera of bacteria, for the samples by platform Illumina (application 16S Metagenomics). Some graphics were obtained by SPSS v 21 (SPSS Inc., Chicago, IL, USA).
Ethics Statement
Studies with human samples (biopsy and brush colorectal samples) were approved by the Ethics Committee of the Balearic Islands (IB3132/16 PI). Data about blood and urine biochemistry markers were proved by the medical researchers, according to the Protection Law of Data Use.
Results
Sensibility and precision of metagenomic method is difficult to demonstrate with a small number of samples or of subjects. However, if the size of fragments obtained as libraries are adequate and the percentage of reads identified is more than 95% (with lower variation coefficient than 3 %); there are some evidences that the results are correct.
Ten subjects with digestive disorders symptoms were analyzed by colonoscopy test, with 32 samples corresponding to different areas of the colon: NEAP (samples of healthy area by biopsy and by brush); UC, CD, Diver and Polyps (samples of healthy area by biopsy and by brush, inflamed area by biopsy and by brush).
Libraries were performed by 16S Amplicon procedure, which generated rRNA fragments with mean size of 604 pb (as indicator of quality control) and an amount of 31 nM by (40 nM estimated by the fluorescent method); and by XT Nextera Metagenomic with a real concentration of 5.31 nM (5 nM estimated) and the size with 1.121 pb, with homogeneous picks detected by the Bioanalyzer technique. Taxonomy was evaluated using 16S Amplicon sequencing application by BaseSpace platform (Illumina), yielding 21.138.542 reads for genera level (a satisfactory percentage of reads identified: 96.65%, variation coefficient: 2.21%, value of cluster density 888 + 6 K/mm2 and 12.11 Gbp). Therefore, the libraries preparation was secure for the next sequencing step. Sensibility observed in our system of samples (for 451 genera) was 30 genera detectable with more than 1% of abundance.
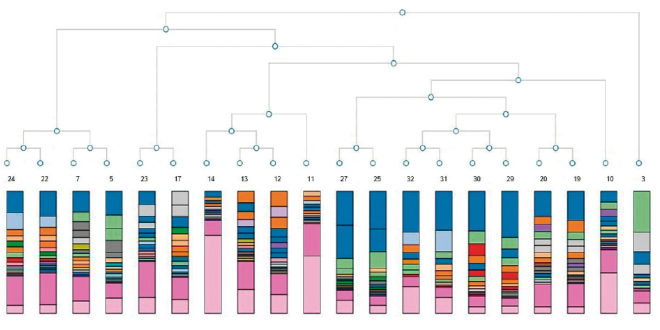
16S Amplicon analysis allowed identify points of similitude and difference by the taxonomy tree, (Figure 1) and PCA (Figure 2), based on abundance of classified operational taxonomic units (OTUs) presented in all samples as differential constellations; in each group.
Figure 1. Taxonomy tree 16S Amplicon analysis.
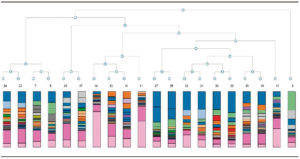
Figure 2. PCA of bacterial diversity by 16S Amplicon analysis.
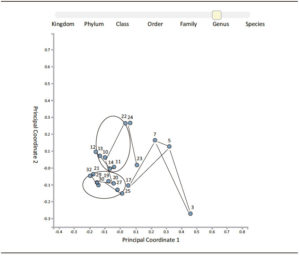
UC samples were outside of CD and Polyps constellations, while samples of NEAP were included inside and in the limit of UC constellation.
Although diverticula pathology is not classified as inflammatory disease, Diver samples were included inside CD constellation.
A possible hypothetic flow of bacterial amount was represented (mucosa-lumen and healthy area-inflamed area) (Figure 3). That proposed a comparative model of bacteria distribution in different areas in the same subject. According to the severity of the disease, bacteria were concentrated in the luminal inflamed area (both P1 and P3). The clinical description for patients showed (Table 1): P1 exhibited in addition polyps, chronic hepatitis; P3 showed a colon sessile polyp (3mm) and a semi-pedunculate 10 mm polyp with an adenomatous appearance and some diverticula in sigma. P2 showed a colon sessile 10 mm polyp without higher dysplasia degree, its bowel bacteria were concentrated in both healthy and inflamed in luminal areas.
Analysis in ulcerative colitis suggested an equal distribution of bacteria, in both areas, for two patients. The inflammation was detected by a colonoscopy test in the reduced inflamed zone of UC1. There were some aphtoid ulcers (with white halo erythematous) and lymphoid hyperplasia associated with ulcerative colitis. That behavior was specific to bacteria concentrated in luminal space (Figure 3).
Figure 3. Distribution of total bacteria in mucosa-lumen and healthy-inflamed area, referred to reads number.
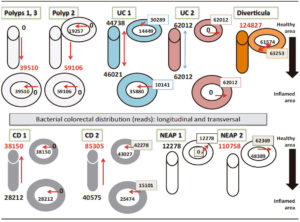
However, UC2 presented an inflammation extended to all studied areas (active colitis in colonic mucosa) by a colonoscopy test (the patient has shown some symptoms with mucus secretion and blood in feces), which exhibited the highest bacterial distribution in all mucosal area and the lumen was not implicated (Figure 3).
Crohn´disease model was particularly noticeable. Both patients had a directional flow out to inflamed areas, so the bacteria amount in the healthy area was double in size (in lumen for CD1 and distributed in both for CD2). Exploration showed CD1 had some starry ulcers with a fibrinous background, upholstered with mucopurulent exudates; there were evidences of preserved mucus secretion and loss of epitheliod surface in some fragments, diffuse inflammation with abundant eosinophils and polymorphonuclears neutrophils, without cryptic bifurcations. CD2 showed an extensive affected mucosal area with some fragments of tubulovillous adenoma; without high grade dysplasia; ileum mucosa and colon mucosa affected with mucus secretion; serrated adenoma sessile not dysplasia; some fragments with distorted cryptic architecture, with crypts of eosinophils, polymorphonuclear, neutrophils; not granulomes or cryptic abscesses, or malignances.
The patient with diverticula had a bacterial diversity pattern similar to CD or included in the constellation of CD (Figure 2); but the bacterial amount was higher distributed in luminal and mucosal areas around the diverticula structure, seen in the flow model (Figure 3).
The NEAP subjects went to digestive medical assistance because both suffered pain associated to fat consumption, with episodes of rectal bleeding, constipation and bowel stools. Both subjects did not show gut inflammatory evidence by colonoscopy. The total bacteria amount in NEAP1 was 4-8 times less than Polyp, UC, CD groups; 10 times less than Diver and it was concentrated in mucosa area without perceptible distribution in luminal portion (Figure 3). Nevertheless; bacterial diver sity in NEAP1 (HABio) conformed to the UC pattern (sample 10, Figure 2); while NEAP2 was outperforming NEAP1 about 10 times higher levels of bacteria distributed in both areas (mucosa and luminal). NEAP2 diversity was in the border of UC (Figure 2).
Therefore, the Illumina platform showed that there were differences among subjects (Table 2). Our results indicated that there were bacterial abundance and diversity differences associated to mucosal areas and lumen, but the areas were in the same constellation, for the same group of patients.
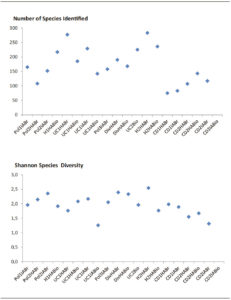
Although there was no correlation between Shannon´s Index and the number of species identified (Pearson´s correlation: p = 0.062); Figure 4 confirmed the major values of bacteria diversity (by Shannon´s Index) had to the major number of species identified for NEAP subjects (H2HABr > H2HABio > H1HABio), containing the major number of genera represented (77 genera) at H2HABr.
Diversity was lowest in sample 14 (UC1 IABio), in CD2 IABr (sample 31) and IABio (sample 32), by Shannon´s Index (1.257; 1.312; 1.239; respectively).
Table 2. Principal bacteria genera found in the samples, by 16S Metagenomics application and XT Nextera Metagenomic (correlation r = 0.7 – 0.83).
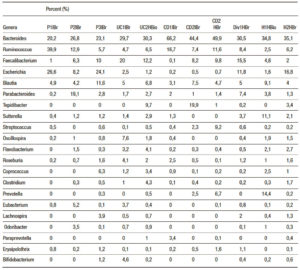
Figure 4. Relationship between number of species identified and Shannon´s Index (by Illumina platform).
MetaPhlAn was applied to analyse XT Nextera Metagenomic products for four samples (Pol2IABr, UC1IABr, H2HABr, CD2HABr), which were carried out in the same sequencing cartridge with 16S Amplicon products, as a quality control criteria. Some sequences could not be read in 16S Amplicon products with appropriate sensitivity in samples 11 and 14 (UC1 HABr and UC1 IABio; pink color in Figure 1); when metagenomic fragments were applied together. Nevertheless, we propose it will be effective to use different sequencing cartridges for each type of sample (amplicon or metagenomic fragment) and the proposed correlation analysis. Table 2 shows the abundant genera in our samples. Bacteroides and Ruminococcus were more representative in CD1IBr, while Faecalibacterium was abundant in UC1IBr. Ruminococcus and Escherichia were evident in P1IBr.
Nutritional quality of the subjects
We elaborated some spreadsheets with the principal groups of foods consumed by the Majorca population (rural and urban population); according to the criteria of Food Composition Spanish Data Base (called BEDCA) for future application. In this work, we used nigh general groups from BEDCA and the criteria recommended by Norte and Ortiz11 to express the nutritional quality as Healthy Eating Index (HEI).
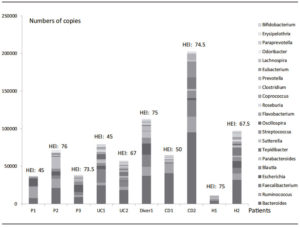
Table 3 was organized including the characteristics of patients and the nutritional pattern. Results confirmed that the participants needed to modify their habitual diet; as increasing the consumption of vegetables, cereals and fruits; decreasing the intake of meat or increasing fish consumption. Figure 5 shows the total content of different genera with HEI.
Table 3. Healthy Eating Index analysis and valuation of nutritional quality.

The lower HEI values were characteristic in one of the patients, in all groups: P1 had higher level of Ruminococcus and Escherichia, UC1 had higher levels of Bacteroides and Faecalibacterium, CD1 had higher levels of Bacteroides and Ruminococcus, and H2 had higher levels of Bacteroides and Escherichia.
The abundance of Prevotella in H1 could be of important significance in the maintenance of higher values of HEI (Table 3, Figure 5).
Diver subject microbiota (HEI: 75) combined some abundance patterns: Bacteroides, Ruminococcus, Faecalibacterium, Escherichia and additionally, Blautia, Parabacteroides and Sutterella.
Discussion
There is not a single cause that explains the origin of bowel inflammatory diseases. Acute outbreaks can be triggered by infections, non-steroidal anti-inflammatory drugs and stress. Internal/external environmental factors in the host and bacteria produce an anomalous inflammatory response that is perpetuated over time producing the disease.12, 13
We took as a reference the results obtained by Lavelle et al.3 who demonstrated the interpersonal variability in luminal and mucosal areas, in gut for communities of bacteria, associated with ulcerative colitis. We could extend those analyses to Crohn´s disease, polyps and patients affected by intestinal diverticula. Our study allowed the establishment of the methodology for collection of biopsy and brush samples, storage, isolation of bacterial DNA and library preparation. Our proportion solid sample: volume of extraction buffer was higher than the proportion used by Lavelle et al,3 because we considered that it was better for the sterile manipulation by the sanitary collector. We processed the total sample which allowed the adequate amount of DNA for the library performances and the specimen was enough for sequencing the amplified and purified rRNA fragments.
We consider that two or three subjects in group were not enough to demonstrate the bacteria “core”. However, there are some studies which have demonstrated there is not general pattern because of the individual difference: … “Each person´s microbiome has its own unique combination”.2 Interpersonal differences make it necessary to study case by case.
We did not consider to NEAP subjects as healthy; although the exploration was normal. In those cases, clinical indications could suggest an irritable bowel syndrome (IBS) without a clear mucosal damage. Some characteristics were detected in both subjects: the bacteria levels and diversity in H1 were lower than H2 (in lumen); and H1 had levels of Escherichia ten times lower than H2 (Table 2).
At the same times, Prevotella could be an important element in H1 for the control of Bacteroides and Escherichia levels. These mechanisms are known as inter-kingdom chemical sensing systems.14 Prevotella genus in rumen strains has been reported that the QS system AI-2 is induced by glucose;15 and traditional diets rich in plant polysaccharides are associated with its prevalence.16
Although diverticula pathology is not considered an inflammatory disease; sometimes, the researchers forget that the fact of changing in distribution and bacterial composition in mucosa, in epithelial barrier and blood flow could induce inflammatory and neuromuscular changes associated to gut diverticula disease.17
In the methodology proposed in this work, we recommend including polyps and diverticula pathologies and subjects with antecedents or predisposition to inflammatory diseases.
In our results, the lowest results of nutritional qualifiers (Table 3) were not in patients more affected, in each group of diseases. But this was a fact that the consumption patterns were not satisfactory. Norte and Ortiz11 obtained a media punctuation of HEI (IASE) as 72.1 (n = 1603), for Balearic Island; so, that was one of those presented higher index of population with a food as little-healthy.
Every time, more institutions are added to the development of tests for gut microbiota characterization as a complementary tool to traditional diagnosis. Some platforms address taxonomic criteria to predict future infections by pathogens, to indicate treatments and recommend diet modifications. The cost of determinations is variable and it depends on the arm of the study. In such way, the platform uBiome (of the cheapest) has set a price for the study of $89 by sample, (GutExplorer) a price of $ 199 for a complete evaluation before and after a treatment (3 samples).
Other platforms have some ranges of price almost $ 99 – 135 and $ 349 – 399 (as American Gut, BIOHM, Day Two, THRYVE, and VIOME); depending of if it includes a nutritional valoration (https:// rawlsmd.com).
We estimated a preliminary cost for the evaluation in concept of research objective by our system of evaluation (58.5 €/sample). It will depend on the complexity of the samples, ability of the collector, the direct and indirect costs. In this way, if this methodology could be applied in our care centres, and establishing the criteria; in term of the trend of bacterial variability and nutritional styles; it would be possible to reduce the costs of the application.
Undoubtedly, the prevention is important because changes in healthy bacteria pattern are associated to epidemics as by Clostridium difficile in United Stated. The cost of a treatment with vancomicine 125 mg, by 10 days, is USD 1779 if the infection is not recidivist.19
The availability of the metagenomic technology as reference will be essential for the design and development of simple, rapid and economic complementary methods with colonoscopy test.
Conclusions
The first step to establish gut microbiota metagenomic technology in our group, allowed us to obtain bacteria genomic libraries by colonoscopy exploration, from patient with bowel diseases; according to Illumina requirements.
Preliminary results will permit to introduce the metagenomic methodology; increasing the subjects number and samples and their nutritional characterization.
Conflict-of-interest statement. No potential conflicts of interest.
Supporting. Department of Innovation, Interior and Justice, Government of Balearic Islands, Spain, for funding a Special Support (AAEE 27/2016).
Acknowledgments. To the memory of our professor Francisco José García-Palmer. We thank the Department of Innovation, Interior and Justice, Government of Balearic Islands, Spain, for funding a Special Support (AAEE 27/2016). We thank the volunteers for their participation. We thank Fundación para el Fomento de la Investigación Sanitaria y Biomédica (FISABIO, Comunidad Valenciana), especially, Dr. Llùcia Martínez Priego and Dr. Giuseppe D’Auria; and Dr. Esther Castillo (Library Preparation Specialist, Illumina®, Spain) who contributed us with their experiences in “Library Curse” April 2016, Valencia, Spain. We thank Fundamental Biology Department and Grup de Metabolisme Energètic i Nutrició, University of Balearic Islands, Spain.
References
- Jandhyala M, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Reddy N. Role of the normal gut microbiota. World J Gastroenterol 2015; 21: 8787-8803.
- Kellman R. The microbiome diet. The scientifically proven way to restore your gut healthy and achieve permanent weight lost. Da Capo Lifelong Books, Boston 2014: 11-12.
- Lavelle A, Lennon G, O’Sullivan O, Docherty N, Balfe A, Maguire A, Mulcahy HE, Doherty G, O’Donoghue D, Hyland J, Ross RP, Coffey JC, Sheahan K, Cotter PD, Shanahan F, Winter DC, O’Connell PR. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 2015; 64: 1553-1561.
- Khanna S, Tosh PK. A clinician’s primer on the role of the microbiome in human health and disease. Mayo Clin Proc 2014; 89: 107-114.
- Guarner F, Malagelada J. «Gut flora in health and disease». Lancet 2003; 361: 512-519.
- Beaugerie L, Petit JC. Antibiotic-associated diarrhoea. Best Practice & Research Clinical Gastroenterology 2004; 18: 337-352.
- Dehingia M, Thangjam devi K, Talukdar NC, Talukdar R, Reddy N, Mande SS, Deka M, Khan MR. Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Scientific Reports 2015; 5: 18563.
- Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Ther Adv Gastroenterol 2013; 6: 295-308.
- Vidal R, Ginard D, Khorrami S, Mora-Ruiz M, Muñoz R, Hermoso M, Díaz S, Cifuentes A, Orfila Alejandro, Roselló-Móra R. Crohn associated microbial communities associated to colonic mucosal biopsies in patients of the western Mediterranean. Systematic and Applied Microbiology 2015; 38: 442-452.
- Spanish Society of Community Nutrition. Spanish Healthy Eating Guide 2004. Available from: http://www.nutricioncomunitaria.org/generica.jsp?tipo=docu&id=3
- Norte A, Ortiz R. Calidad de la dieta española según el índice de alimentación saludable. Nutr Hosp 2011; 26: 330-336.
- Qin L, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464: 59-65.
- Medina E. Enfermedad inflamatoria intestinal (I): clasificación, etiología y clínica. A Pediatr Contin 2013; 11: 59-67.
- Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fructose sensing regulates bacterial intestinal colonization. Nature 2012; 492: 113-117.
- Gorenc G, Lukas F, Avgustin G. Examination of ai-2 quorum sensing system in Prevotella bryantii and Prevotella ruminicola-like strains by using bioluminiscence assay. Acta Agriculturae Slovenica 2007; 90: 107-113.
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, BaldassanoR, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011; 334: 105-108.
- Quigley EM. Gut microbiota, inflammation and symptomatic diverticular disease. New Insights into an Old and Neglected Disorder. J Gastrointestin Liver Dis 2010; 19: 127-129.
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 2007; 104: 13780-13785.
- Nelson R, Suda KJ, Evans CT. Antibiotic treatment for associated diarrhoea in adults. Cochrane Database of Systematic Reviews 2017; 3: CD004610.
Correspondencia: Ileana Martínez-Cabrera
Grupo Metabolismo Energético y Nutrición. Carretera de Valldemossa, Palma, 07122, Ille Balears. Universitat de les Illes Balears, España.
Tel.: +34 622075746
Correo electrónico: ileanamart@gmail.com
Acta Gastroenterol Latinoam 2019;49(2): 96-109
 Revista ACTA Órgano Oficial de SAGE
Revista ACTA Órgano Oficial de SAGE