Jéssica Tonin Ferrari1,2* ID· Gabriel Tayguara Silveira Guerreiro1,2* ID · Larisse Longo1,2 ID · Themis Reverbel da Silveira2 ID· Carlos Thadeu Schmidt Cerski1,3 ID · Erica Tozawa3 ID · Cláudia P Oliveira4 ID· Mário Reis Álvares-da-Silva1,2,5,6 ID· Carolina Uribe-Cruz1,2,7 ID
1 Graduate Program in Gastroenterology and Hepatology, Universidade Federal do Rio Grande do Sul, Porto Alegre. Rio Grande do Sul, Brazil.
2 Experimental Laboratory of Hepatology and Gastroenterology, Center for Experimental Research, Hospital de Clínicas de Porto Alegre, Porto Alegre. Rio Grande do Sul, Brazil.
3 Unit of Surgical Pathology, Hospital de Clínicas de Porto Alegre, Porto Alegre. Rio Grande do Sul, Brazil.
4 Department of Gastroenterology (LIM07), Faculdade de Medicina da Universidade de São Paulo. São Paulo, Brazil.
5 Division of Gastroenterology, Hospital de Clínicas de Porto Alegre, Porto Alegre. Rio Grande do Sul, Brazil.
6 Conselho Nacional de Desenvolvimento Científico e Tecnológico, CNPq Researcher.
7 Universidad Católica de las Misiones, Posadas. Misiones, Argentina.
* Jéssica Tonin Ferrari and Gabriel Tayguara Silveira Guerreiro contributed equally to the writing of this article.
Acta Gastroenterol Latinoam 2023;53(3):265-282
Received: 10/06/2023 / Accepted: 04/09/2023 / Published online: 29/09/2023 /
https://doi.org/10.52787/agl.v53i3.329
Summary
Aim. To evaluate the effects of rifaximin through microbiota modulation in a model of hepatocellular carcinoma secondary to non-alcoholic fatty liver disease. Methods. Three groups of
8 adult male Sprague-Dawley rats each were divided as follows: the HCC group: rats fed a high-fat and choline-deficient diet plus diethylnitrosamine as a carcinogen, the hepatocellular carcinoma treated group: rats fed a high-fat and choline-deficient diet plus diethylnitrosamine and treated with rifaximin and the control group: animals fed standard diet and water. The rats were euthanized after 16 weeks. We performed analyses of liver pathology for non-alcoholic fatty liver disease severity and cancer grading, gene expression in intestinal and hepatic tissues and fecal microbiota. Results. All animals in the hepatocellular carcinoma group had non-alcoholic fatty liver disease and developed hepatocellular carcinoma lesions. Rifaximin animals showed less intense non-alcoholic fatty liver disease (assessed by non-alcoholic fatty liver disease activity score [NAS]) compared to the hepatocellular carcinoma group. Both the hepatocellular carcinoma and hepatocellular carcinoma + rifaximin groups showed areas of fibrosis as assessed by picrosirius red. Three animals in the rifaximin group did not develop cancerous lesions. Gut microbiota analyses revealed differences in diversity and composition in the control group vs hepatocellular carcinoma and rifaximin groups. Twelve differentially abundant genera were identified between the hepatocellular carcinoma and rifaximin groups. In the rifaximin group, gene expression of intestinal tight junctions decreased. Conclusions. In a rodent model of non-alcoholic fatty liver disease-related hepatocellular carcinoma, rifaximin reduces the histological severity of non-alcoholic fatty liver disease and the occurrence of hepatocellular carcinoma, probably by modulating the gut microbiota independently of markers of intestinal permeability.
Keywords. Gut microbiota, hepatocellular carcinoma, non-alcoholic fatty liver disease, rifaximin.
Beneficio potencial de la rifaximina en la prevención del carcinoma hepatocelular mediante la modulación de la microbiota en un modelo experimental de enfermedad por hígado graso no alcohólico
Resumen
Objetivo. Evaluar los efectos de la rifaximina mediante la modulación de la microbiota en un modelo de carcinoma hepatocelular secundario a enfermedad por hígado graso no alcohólico. Métodos. Se dividieron tres grupos de 8 ratas Sprague-Dawley macho adultas cada uno de la siguiente manera: el grupo carcinoma hepatocelular: ratas alimentadas con una dieta alta en grasas y deficiente en colina más dietilnitrosamina como carcinógeno; el grupo tratado con carcinoma hepatocelular: ratas alimentadas con una dieta alta en grasas y deficiente en colina más dietilnitrosamina y tratadas con rifaximina y el grupo control: animales alimentados con una dieta estándar y agua. Las ratas fueron sometidas a eutanasia a las 16 semanas. Se realizaron análisis de la patología hepática para determinar la gravedad de la enfermedad por hígado graso no alcohólico y la clasificación del cáncer, la expresión génica en tejidos intestinales y hepáticos y la microbiota fecal. Resultados. Todos los animales del grupo de carcinoma hepatocelular tenían enfermedad por hígado graso no alcohólico y desarrollaron lesiones de carcinoma hepatocelular. Los animales del grupo con rifaximina mostraron una enfermedad por hígado graso no alcohólico menos intensa (evaluada por el puntaje de actividad de la enfermedad por hígado graso no alcohólico NAS]) en comparación con el grupo carcinoma hepatocelular. Los grupos carcinoma hepatocelular y carcinoma hepatocelular + rifaximina mostraron áreas de fibrosis evaluadas con rojo picrosirio. Tres animales del grupo con rifaximina no desarrollaron lesiones cancerosas. Los análisis de la microbiota intestinal mostraron diferencias en la diversidad y composición de los grupos control vs carcinoma hepatocelular y rifaximina. Se identificaron 12 géneros diferencialmente abundantes entre los grupos carcinoma hepatocelular y rifaximina. En el grupo con rifaximina disminuyó la expresión génica de las uniones estrechas intestinales. Conclusiones. En un modelo de roedores de carcinoma hepatocelular relacionado con enfermedad por hígado graso no alcohólico, la rifaximina disminuye la gravedad histológica de la enfermedad por hígado graso no alcohólico y la aparición de carcinoma hepatocelular, probablemente mediante la modulación de la microbiota intestinal independientemente de los marcadores de permeabilidad intestinal.
Palabras claves. Microbiota intestinal, carcinoma hepatocelular, enfermedad del hígado graso no alcohólico, rifaximina.
Abbreviations
ALT: Alanine aminotransferase.
AST: Aspartate aminotransferase.
Cldn: Claudin.
CON: Control group.
DEN: Diethylnitrosamine.
JAM/FR11: Junctional adhesion molecule.
HDL: High-density lipoprotein.
HFCD: High-fat and choline-deficient.
IL: Interleukin.
LEfSe: Linear discriminant effect size.
LDA: Linear discriminant analysis.
LDL: Low-density lipoprotein.
NAFLD: Nonalcoholic fatty liver disease.
NAS: NAFLD activity score.
NASH: Nonalcoholic steatohepatitis.
HCC: Hepatocellular carcinoma.
Ocln: Occludin.
OTUs: taxonomic units.
RIF: Group treated with rifaximin.
TNF: Tumor necrosis factor.
TJP1/ZO-1: Tight junction protein-1.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is one of the most common forms of chronic liver disease in western developed countries.1 Non-alcoholic steatohepatitis (NASH) represents the inflammatory form of the disease, which can progress to cirrhosis and hepatocellular carcinoma (HCC).2,3 HCC is the fifth most common malignant neoplasm in the world and is considered the most common form of liver cancer, accounting for 70-85% of the primary liver tumors.4,5 Although several therapies are currently available, new therapeutic approaches to prevent the development and progression of this disease are still needed.
Dysbiosis is a disorder in the structure or function of the microbiota resulting in an abnormal balance of commensal and pathogenic bacterial species. This imbalance is present in NAFLD and alters several processes that influence the progression of chronic liver disease, such as inflammation, fibrogenesis, regeneration and immunity, thereby affecting the development of HCC.6-11 Various forms of interventions can be used to address gut dysbiosis, including prebiotics, probiotics, symbiotics or even antibiotics. In fact, antibiotics can modulate gut microbiota, improve endotoxemia and prevent liver progression and decompensation.12-14 Rifaximin is a non-absorbable, broad-spectrum, oral semi-synthetic antibiotic that inhibits bacterial RNA synthesis by binding to the β-subunit of bacterial DNA-dependent RNA polymerase.15 By altering the gut microbiota, it modulates its function and the levels of inflammatory mediators. Rifaximin acts locally in the gastrointestinal tract and has low systemic absorption with minimal adverse effects. It has been used in the treatment of various gastrointestinal and liver conditions,16 but few studies have evaluated its role in NAFLD17-19 and, to our knowledge, none have investigated it in NAFLD-related HCC. Given the beneficial effects of rifaximin in preventing dysbiosis in liver disease progression, this study aims to evaluate the effect of rifaximin on gut microbiota in a murine model of NAFLD-related HCC.
Methods
Animals
In this study, we used 24 adult male Sprague-Dawley rats, weighing 250-400 g, obtained from the Animal Experimentation Unit of the Hospital de Clínicas de Porto Alegre. The animals were housed in boxes of up to three rats, with access to food and water ad libitum, under a
12 hours light/dark cycle and at a temperature of 22 ± 1°C. The experimental protocol was designed to minimize pain or discomfort to the animals. All procedures were in accordance with the current Brazilian legislation and were approved by the Ethical Committee for the Use of Animals under number 17-0087. In case of death before euthanasia, the animal was excluded from the study.
Experimental design, model of hepatic carcinogenesis secondary to NAFLD and treatment with rifaximin
Animals were randomly assigned according to their weight so that all groups started the experiment homogeneously and were divided into three groups (n = 8/group): in the HCC group, rats received a high-fat and choline-deficient (HFCD) diet (35% total fat, enriched with 54% trans fatty acids) (Rhoster Ltd., Brazil) together with 135 mg/L of diethylnitrosamine (DEN; Sigma-Aldrich, United States) in the drinking water for 16 weeks.20 In the RIF group, rats received the HFCD diet plus DEN, and rifaximin (Sigma-Aldrich) at a dose of 50 mg/kg/day by daily gavage, diluted in distilled water, as previously published, starting at week 5.14 In the control group (CON), rats received standard chow (Nuvilab CR-1, Brazil) and water free of DEN. Animals in the CON and HCC groups received daily gavage with distilled water in order to undergo the same stress conditions as those in the RIF group. All animals were weighed weekly throughout the experiment and were euthanized after 16 weeks.
Euthanasia and analysis of biochemical parameters
After 16 weeks, all animals were fasted for eight hours, anesthetized with 5% isoflurane (BioChimico, Brazil) in 100% oxygen and euthanized by cardiac exsanguination. After blood collection, the serum was separated for biochemical analyses of aspartate aminotransferase (AST), alanine aminotransferase (ALT), total cholesterol, triglycerides, high-density lipoprotein (HDL), and low-density lipoprotein (LDL), which were performed in routine automated equipment at the Hospital de Clínicas de Porto Alegre (Siemens Advia 1800 Chemistry System). After euthanasia, the liver, intestine, and stool samples were collected and stored appropriately for further analyses.
Anatomopathological evaluation
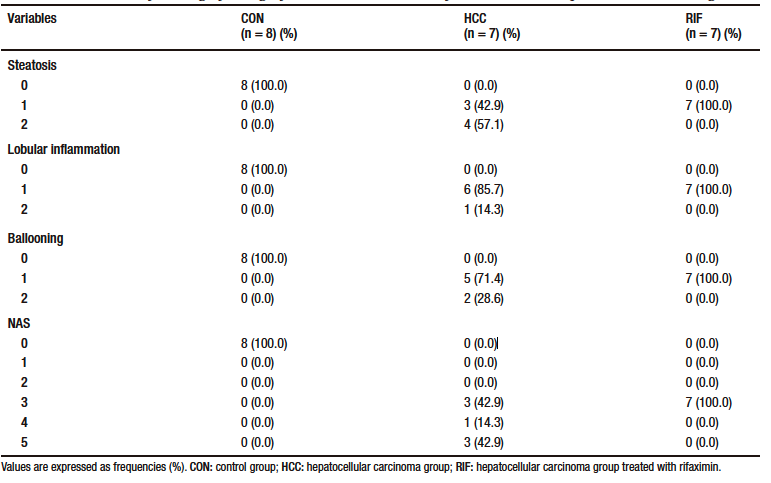
After euthanasia, a portion of the liver tissue was preserver in 10% formaldehyde (Synth, Brazil) and sectioned at 3-μm. Sections were stained with hematoxylin & eosin and picrosirius red. Slides were evaluated by two blinded experienced pathologists. Slides were photographed using an optical microscope (Olympus BX51). Histological variables were graded using a modified Kleiner system: steatosis (0-3), lobular inflammation (0-3), hepatocellular ballooning (0-2) and fibrosis (0-4).21 After evaluation, the NAFLD activity score (NAS) was determined, where scores of 1 or 2 corresponded to the absence of NASH, scores of 3 or 4 corresponded to a probable diagnosis and scores greater than or equal to 5 corresponded to a definitive diagnosis of NASH. The degree of fibrosis was assessed using picrosirius red stained slides and cancerous lesions were graded according to the Edmondson & Steiner classification (1 to 4).22
Gene expression of inflammatory cytokines in the liver and tight junctions in the intestine
After euthanasia, the liver and small intestine were collected and total RNA was extracted with TRIzol (Invitrogen, United States); RNA was later converted to cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, United States). qRT-PCR with TaqMan assay (Applied Biosystems, USA) was used to assess the gene expression of hepatic inflammatory cytokines: interleukin (Il)-6, Il10, Il1b, and tumor necrosis factor-α (Tnfa) and intestinal tight junction proteins: occludin (Ocln), claudin (Cldn)-2, Cldn4, zona occludens (Zo1) and junctional adhesion molecule (JAM/F11r). The Actb and Gapdh genes were used as normalisers (Supplementary Table-1). Values were calculated as a percentage of the normalising gene.
DNA extraction and 16S rRNA sequencing
The colon was dissected, and stool samples were collected and transferred to a tube. Procedures and materials were sterile to avoid contamination. Bacterial DNA was isolated from the stool samples using the QIAamp fast DNA stool mini kit (Qiagen, United States), according the manufacturer’s instructions. The hypervariable V4 region of the 16S rRNA gene was amplified by PCR using the following primer pair: 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). To pool different samples in the same reaction, we used the primer fusion method and each sample had a unique barcode attached to the corresponding PCR product. The purified products were subjected to emulsion PCR using the Ion PGM™ Hi-Q™ View OT2 kit (Thermo Fisher Scientific, United States), and the resulting enriched beads were sequenced on an NGS machine (Ion Torrent PGM, Life Technologies, United States) using the Ion PGM™ Hi-Q™ View Sequencing kit (Thermo Fisher Scientific). Finally, the corresponding bioinformatic analyses were performed (online resource). Additional information on bioinformatics analyses (processing of 16S rRNA reads for downstream analyses) is described in the supplementary material.
Statistical analyses
Normality was tested for all variables using the Shapiro-Wilk test and histograms. For symmetric variables (described by means ± standard deviations), we used the ANOVA followed by the Tukey test, and for asymmetric variables (described by medians and minimum–maximum values), we used the Kruskal-Wallis test followed by the Dunn test. The chi-squared test, supplemented by an analysis of adjusted residuals, was used to obtain percentage data. Analyses were performed using the PASW Statistics package, version 18.0 (SPSS Inc, Chicago, USA). The p value was considered statistically significant if ≤ 0.05.
Alpha diversity was assessed using species richness (Chao1) and species diversity (Shannon) indices. For an overall comparison of significant differences between bacterial communities (i.e., beta diversity), Principal Coordinates Analysis (PCoA) was performed. For each pair of samples, a matrix was calculated using phylogenetic (weighted Unifrac) and non-phylogenetic (Bray-Curtis dissimilarity) metrics. Distances were transformed into points in space, with the number of dimensions being one less than the number of samples. To obtain statistical confidence in the sample grouping observed in the PCoA, a multivariate analysis of similarities (ANOSIM) test was performed on the distance matrix. To compare additional differences between microbial communities, we used clustering methods based on Bray-Curtis dissimilarity. The hierarchical clustering results were visualised using heat maps and dendrograms. We used the Linear Discriminant Effect Size (LEfSe) method to detect potential taxa biomarkers.23 The algorithm performs a non-parametric factorial Kruskal-Wallis sum rank test and a linear discriminant analysis (LDA) to identify significantly different features between taxa and estimate the effect size of the difference. Differences were considered significant for a logarithmic LDA score threshold of ± 2.0 and a p-value < 0.05 after multiple hypotheses testing using the Benjamini–Hochberg method.
Results
At 15th week, one animal died in each of the HCC and RIF groups. These animals were excluded from the study. No deaths were reported in the CON group during the experimental period. There was no statistically significant difference in the animals’ delta weight (Supplementary Table-2).
Biochemical parameters and gene expression
Liver enzymes and lipid profiles are described in Supplementary Table-3. AST, total cholesterol and LDL levels were significantly higher in the HCC group compared to the CON group (p = 0.001).
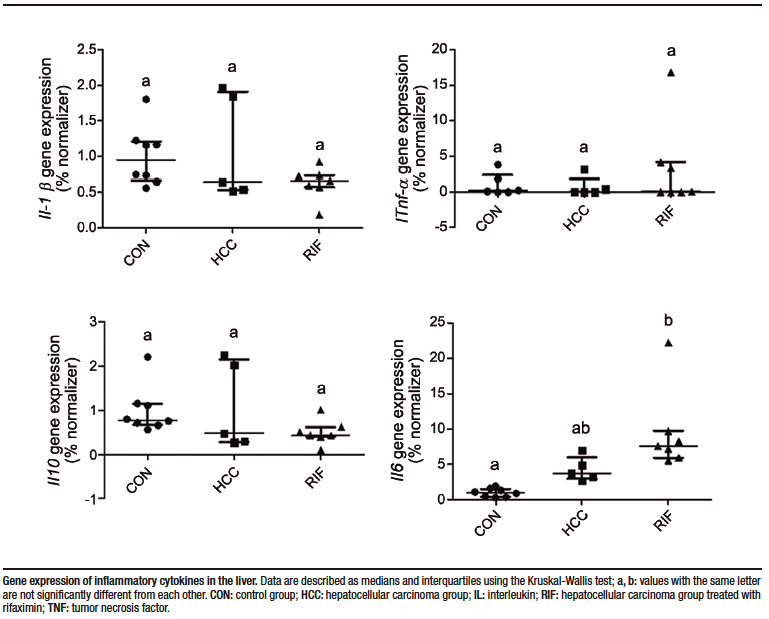
Hepatic gene expression of pro- and anti-inflammatory cytokines were evaluated. Tnfa, Il10, and IL1b showed no differences in expression between groups; however, Il6 expression was significantly increased in the RIF group when compared to the CON group (p = 0.001) (Supplementary Figure 1).
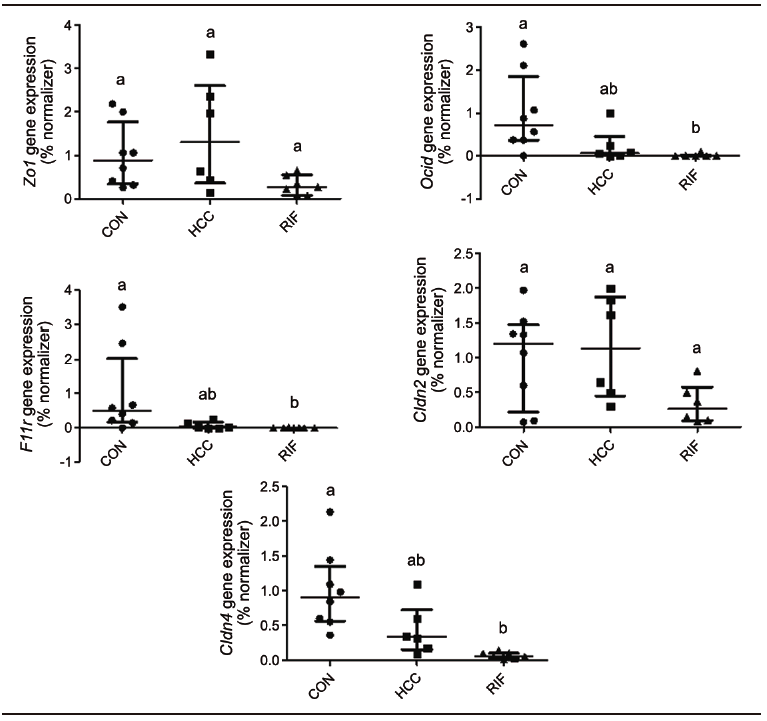
To understand the effect of rifaximin on intestinal permeability, we evaluated the expression of tight junction proteins in the small intestine (Supplementary Figure 2). There were no differences between groups for Zo1/Tjp1 and Cldn2. Ocln, JAM/F11r, and Cldn4 showed lower expression levels in the RIF group when compared to the CON group, indicating a possible effect on intestinal permeability (p = 0.002, p = 0.005, and p = 0.001, respectively), but no differences in the expression of these genes were found between the HCC and RIF groups.
Rifaximin treatment may influence steatohepatitis
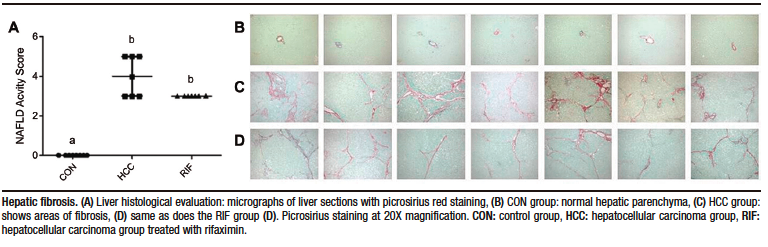
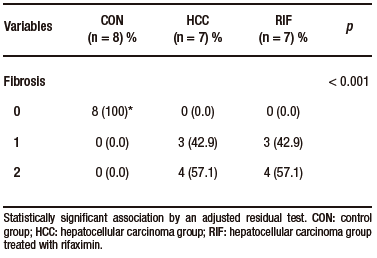
None of the control animals showed features of liver injury, as shown in Table-1. With regard to steatosis, animals in the HCC group had a significantly higher fat content than those in the RIF group. The same happened with hepatocyte ballooning. The HCC group had a NAS between three and five; these scores indicate probable and definitive steatohepatitis, respectively. However, all rifaximin-treated animals remained in grade 3, indicating a probable diagnosis of steatohepatitis, suggesting a moderate efficacy of rifaximin treatment (Table 1). When assessing fibrosis by picrosirius red staining, we observed that both the HCC and RIF groups had areas of fibrosis that were different from the CON group (Supplementary Figure 3). In both the HCC and RIF groups, animals ranged equally from grades 1 to 2. As expected, the CON group was grade 0 (Supplementary Table 4).
Table 1. Distribution of histologic findings of steatosis scores, lobular inflammation, and hepatocellular ballooning
Rifaximin treatment moderately prevents hepatocellular carcinoma
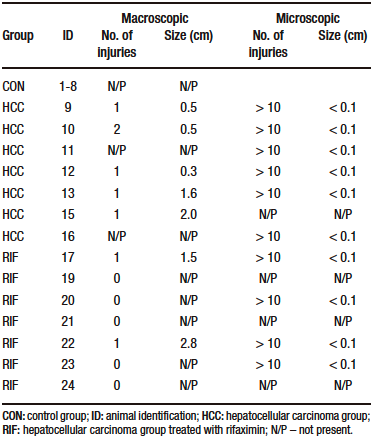
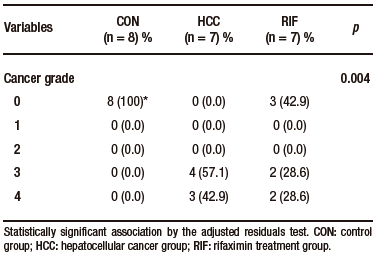
Macroscopically, all animals in the HCC group developed liver tumors that were larger than those in the RIF group. As expected, the control animals had normal livers (Figure 1A-C). Histologic analyses showed that all animals in the HCC group had an altered parenchyma, while treatment with rifaximin appeared to reduce the degree of the lesion. No evidence of hepatic tumor injury was observed in the CON group (Figure 1D-F). Liver lesions are described macroscopically and microscopically in Table 2. When tumors were graded according to the Edmondson-Steiner score (Figure 1G), we observed that all animals in the HCC group had grades three and four, corresponding to poorly differentiated and undifferentiated cancer, respectively. In the RIF group, some animals were also classified as grades three and four, but three animals had no evidence of cancer (Supplementary Table 5).
Figure 1. Hepatocellular carcinoma analysis
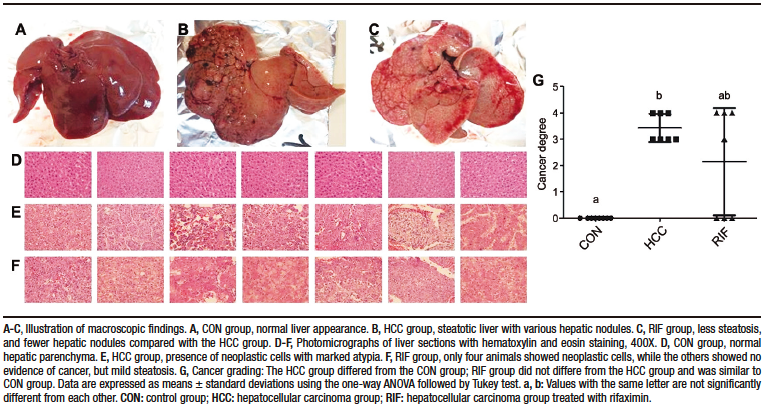
Table 2. Liver injuries description
Gut microbiota
In terms of alpha diversity, no differences were observed between groups with respect to the Shannon index (Figure 2A); however, they differed when considering the Chao1 index, with the RIF group having the lowest diversity and the CON group having the highest diversity (p ≤ 0.01) (Figure 2B). To explore the global differences in gut bacterial communities between groups, a PCoA was performed on operational taxonomic units (OTU)-based Bray-Curtis distances. By examining the PCoA ordination plot, we observed that the structural pattern of the gut microbiota in the HCC and RIF groups was clearly different from that in the CON group (ANOSIM, p < 0.001) (Figure 2C).
Figure 2. Changes in gut microbiota in CON, HCC, and RIF groups
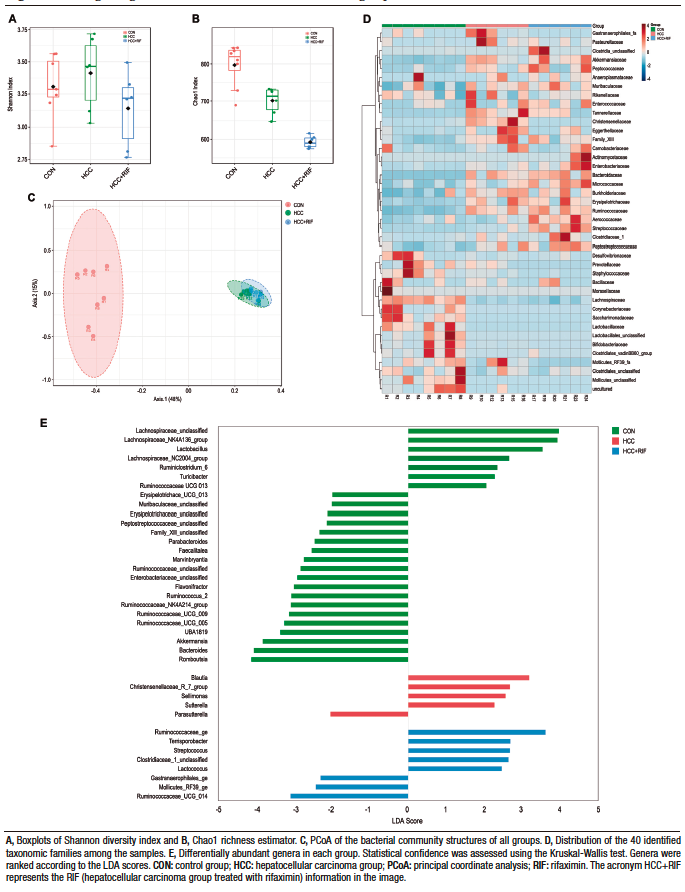
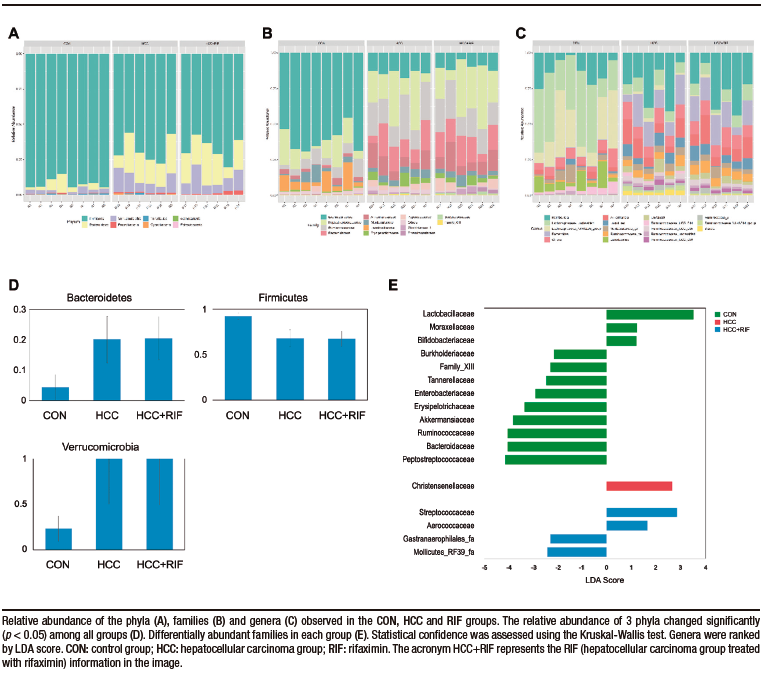
Taxonomic analysis of the bacterial communities identified 1506 bacterial taxa (OTUs), belonging to 124 genera, 40 families, and 8 phyla. Firmicutes (76%), Bacteroidetes (15%) and Verrucomicrobia (7%) were the most common phyla in all samples (Supplementary Figure 4A). These three phyla were also differentially abundant between groups, as observed in Supplementary Figure 4D. Other phyla had less than 1% abundance each. The most abundant families were Lachnospiraceae (28%), Peptostreptococcaceae (21%), Ruminococcaceae (15%), Bacteroidaceae (10%), and Akkermansiaceae (7%). These five families represented 83% of all observed taxa (Supplementary Figure 4B). The distribution of these taxa showed interesting features (Figure 2D), where the most prevalent families in the CON group were less prevalent (or absent) in the HCC and RIF groups, and the most prevalent families in the HCC and RIF groups were less prevalent (or absent) in the CON group. Their differential abundance is showed in Supplementary Figure 4E. The 10 most abundant genera, with a relative abundance greater than 2%, represented 78% of all taxa observed in this study and can be considered a core microbiome (Supplementary Figure 4C). To confirm both statistical and biological taxonomic differences at the genus level between groups, the LEfSe algorithm was used with a logarithmic LDA score cutoff ≥ 2.0. In total, the differential abundance analysis identified 39 differentially abundant genera (Figure 2E), of which 26 were associated with the CON group, five with the HCC group, and eight were differentially abundant in the RIF group. The distribution of all 124 genera among groups is shown in Supplementary Figure 5.
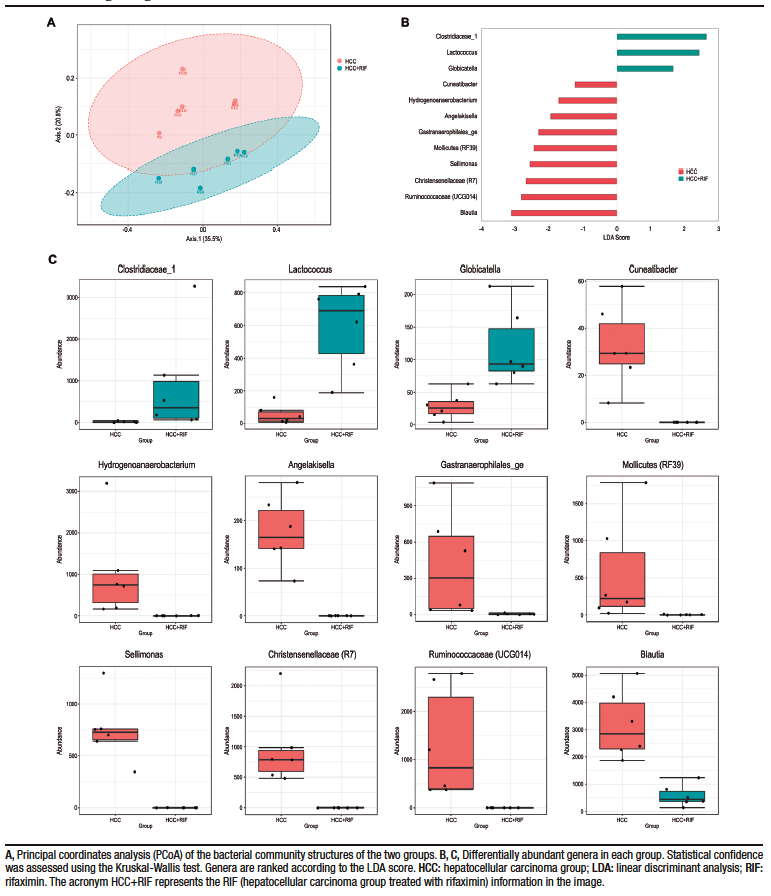
To better understand the differences between the HCC and RIF groups, a PCoA was performed. The two identified clusters are clearly distinct, as observed in Figure 3A, and this pattern was previously overshadowed by the major differences when comparing these groups with the CON group. A specific LEfSe analysis between the HCC and RIF groups revealed 12 differentially abundant genera (Figure 3B), whose profiles are highlighted in Figure 3C.
Figure 3. Changes in gut microbiota between HCC and RIF
Discussion
In this murine model of HCC simulating a western diet associated with DEN, rifaximin demonstrated a potential beneficial effect by reducing the severity of NAFLD and preventing the occurrence of HCC in three out of seven animals, probably due to its effect on the gut microbiota, which in turn would alter certain mechanisms and generate hepatoprotection.
Recent studies have shown that rifaximin is safe and moderately effective in patients with NASH,17-19 but there are no data on its use in HCC. In this regard, we demonstrated a lower degree of steatosis, lobular inflammation, and hepatocellular ballooning in the animals receiving rifaximin; however, further research is needed. The main effect of rifaximin was on steatosis, with a direct influence on NAS; this score was intermediate in all animals that received rifaximin, while more than 50% of animals in the HCC group had higher scores. These findings support the hepatoprotective potential of rifaximin in patients, as do other ongoing clinical trials, such as the randomized controlled phase II LIVERHOPE trial testing the effect of rifaximin in combination with simvastatin.29 Gut microbiota produce a wide range of bioactive molecules from diet, establishing an important role in physiological and pathological conditions, as well as being closely associated with inflammatory diseases and NAFLD and HCC through the gut-liver axis25 which may explain the results of this study.
There is a close relationship between dysbiosis and leaky gut; dysbiosis causes the intestinal barrier to be more permeable, whereas the leaky gut allows bacterial metabolites and microbiota-associated molecular patterns to translocate and reach the liver,26 enabling the progression of NAFLD and HCC.27,28 To elucidate the mechanism involved in the effect of rifaximin, we evaluated the expression levels of tight junction proteins in the intestine and the gut microbiota. In this study, rifaximin did not alter the gene expression of tight junctions, probably through other mechanisms.
When evaluating the gut microbiota, we observed that the HCC group had a different diversity and composition when compared to the CON group. The HCC group showed a decrease in alpha diversity (calculated by Chao1) and a different pattern in beta diversity compared to healthy animals (CON group). These patterns leading to NAFLD and HCC influence the composition and diversity of the gut microbiota.6-10,30,31 Some mechanisms by which the microbiota contributes to the development of HCC are already known.26 Alterations in bile acid metabolism may be associated with the development of HCC, through mechanisms such as metabolic dysregulation, hepatic inflammation, oxidative stress, fibrosis, apoptosis and hyperproliferation, thereby supporting tumorigenesis.32 In dysbiosis, this can be impaired by modulating bile acid metabolism. This altered microbiota could increase deoxycholic acid levels. Enterohepatic circulation of deoxycholic acid induces inflammatory and tumor-promoting factors in the liver, thereby facilitating HCC development in mice.33 Another important mechanism is through lipopolysaccharide, which is a cell wall component of Gram-negative bacteria, which induces inflammation via toll-like receptor-4. This receptor has been shown to mediate hepatic carcinogenesis via resident liver cells such as hepatic stellate cells, macrophages or hepatocytes. In addition to contributing to a chronic inflammatory state, toll-like receptor-4 promotes the development of liver fibrosis and upregulates the expression of epiregulin, a potent HCC-promoter in hepatic stellate cells.14 Although our research group did not perform molecular evaluations of these markers, we suggest that the effects observed in the rifaximin-treated group may result from its action on these inflammatory and hepatic fibrogenic mechanisms.
Increases in the abundance of the phylum Bacteroidetes and the families Bacteroidaceae and Ruminococcaceae have also been found in patients with chronic liver disease in other studies,9,34 suggesting a possible role in the progression of chronic liver disease and, possibly indirectly, in hepatocarcinogenesis. Increases in Ruminococcaceae and Bacteroidaceae were independently associated with the presence of fibrosis and NASH, respectively.9 Possible mechanisms linking the presence of Bacteroidaceae with NASH promotion could be the increase in deoxycholic acid, which is associated with apoptosis induction in rat livers35 and the negative correlation of this family with short-chain fatty acid and amino acid levels,36 thus contributing to NASH promotion. The decrease in Lachnospiraceae in the HCC group, when compared to the CON group, is common in cirrhotic patients and is related to the worsening of chronic liver disease.37,38 Interestingly, we observed an increase in Akkermansiaceae in the HCC group, a family that was recently implicated in hepatoprotection.39,40
Antibiotic therapy in liver disease has been shown to be beneficial in experimental models of NAFLD,41,42 NASH,43,44 and HCC.14 To better understand our results, we compared the gut microbiota between the HCC and RIF groups. When alpha diversity was assessed using the Shannon index (a calculation that takes into account the presence and absence of OTUs), there was no difference between the groups. When evaluated by Chao1 (a calculation that considers the presence and absence of OTUs and is more sensitive to rare OTUs), the RIF group had less alpha diversity than the HCC group. This suggests that rifaximin does not alter the overall diversity of the microbiota, but it may specifically modulate less abundant taxa. The PCoA confirms these findings, where structural differences in the microbiome (i.e., beta diversity) were observed between the HCC and RIF groups. Some studies have suggested that rifaximin may positively modulate the gut microbiota in various gastrointestinal conditions,45,46 and this particular feature may differentiate rifaximin from other systemic antibiotics.47 The LEfSe analysis showed a clear differential abundance between groups at the genus level, with 12 differently abundant genera; these have already been described in other clinical or experimental conditions, but only Blautia has been previously described in detail in HCC and NASH patients.
In this study, we observed that the animals that received rifaximin had an increase in the abundance of the genera Clostridiaceae_1, Globicatella and Lactococcus, and a decrease in the abundance of the genera Cuneatibacter, Hydrogenoanaerobacterium, Angelakisella, Gastranaerophilales_ge, Mollicutes (RF39), Sellimonas, Christensenellaceae (R7), Ruminococcaceae (UCG014), and Blautia. In a study by Bajaj et al, the decompensated cirrhosis group treated with rifaximin had fewer hospitalizations than the lactulose-treated group, and the Lactococcus genus has been described as being associated with fewer hospitalization in cirrhotic patients.48 This genus has also been negatively associated with prokineticin-2, a cytokine associated with inflammation of the gastrointestinal tract, in rats subjected to the Lieber-DeCarli model of alcoholic liver disease.49 A systematic review with meta-analysis found no difference in the abundance of the genus Blautia in patients with NAFLD compared to healthy controls.50 Regarding the other genera, although little information is available, as they were differentially abundant treated or not with rifaximin, they indicate that they could be the subject of further study. As mentioned above, there is evidence that changes in the composition of the gut microbiota play a fundamental rol in the progression of NAFLD and the development of HCC. In this study, we suggest that RIF treatment could mitigate the histopathologic severity of NAFLD and prevent HCC development, likely through the modulation of the gut microbiota. To our knowledge, there are no studies in the literature evaluating the effect of RIF treatment on the composition of the gut microbiota in HCC secondary to NAFLD, which complicates the discussion of the data obtained. In addition, we emphasize that RIF could potentially alter intestinal microbial interactions with metabolites without a significant effect on gut composition or abundance per se. In this regard, further studies are needed for a more comprehensive evaluation of the mechanisms involved in this process.
This is the first article to help elucidate the role of rifaximin in the treatment of NAFLD-HCC through modulation of the gut microbiota. However, it has some deficiencies that need to be emphasized. First, the NAFLD-HCC is a tumor with specific molecular characteristics compared to other etiologies of HCC, second, the microbiota was only assessed at the end of the experiment, third, the number of animals was small. If it were larger could generate more statistical power to the study, reducing the possible bias of model heterogeneity. Nevertheless, we studied the NAFLD Activity Score (NAS) to better understand the underlying disease evolution in the groups. In fact, we consider that the experimental model used in our study, although a mixed model with diet and a carcinogen to induce complex diseases, which could lead to possible heterogeneity, was efficient and replicable with respect to previously published studies.20,24 In conclusion, in this rodent model of NAFLD-HCC, rifaximin may have beneficial effects by reducing the histologic severity of NAFLD and the occurrence of HCC, probably by modulating the gut microbiota independently of markers of intestinal permeability. However, further studies are needed to demonstrate the protective effect of rifaximin and the mechanism by which rifaximin might have a beneficial effect.
Intellectual Property. The authors declare that the data, figures, and tables that appear in this article are original and were made in their belonging institutions.
Funding. This study was supported by the following Brazilian funding agencies: Research Incentive Fund from Hospital de Clínicas de Porto Alegre (Dr. M.R. Álvares-da-Silva, grant 2017-0087); National Council for Scientific and Technological Development, CNPq (M.R. Alvares-da-Silva); and Coordination for the Improvement of Higher Education Personnel, CAPES/PNPD.
Conflict of Interest. The authors declare that they have no conflicts of interest in relation to this article.
Copyright
© 2023 Acta Gastroenterológica latinoamericana. This is an open-access article released under the terms of the Creative Commons Attribution (CC BY-NC-SA 4.0) license, which allows non-commercial use, distribution, and reproduction, provided the original author and source are acknowledged.
Cite this article as: Tonin Ferrari J, Tayguara Silveira Guerreiro G, Longo L et al. Potential Beneficial Effect of Rifaximin in thePrevention of Hepatocellular Carcinoma through the Modulation of the Microbiota in an Experimental Model of Non-alcoholic Fatty Liver Disease. Acta Gastroenterol Latinoam. 2023;53(3):265-282. https://doi.org/10.52787/agl.v53i3.329
References
- Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol [Internet]. 2018 Jan 20;15(1):11-20. Available from: http://www.nature.com/articles/nrgastro.2017.109
- Than NN, Newsome PN. A concise review of non-alcoholic fatty liver disease. Atherosclerosis [Internet]. 2015 Mar;239(1):192-202. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0021915015000301
- Lewis JR, Mohanty SR. Nonalcoholic fatty liver disease: A review and update. Dig Dis Sci [Internet]. 2010 Mar 26;55(3):560-78. Available from: http://link.springer.com/10.1007/s10620-009-1081-0
- Gomes MA, Priolli DG, Tralhão JG, Botelho MF. Hepatocellular carcinoma: Epidemiology, biology, diagnosis, and therapies. Rev Assoc Med Bras [Internet]. 2013 Sep;59(5):514-24. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0104423013001462
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin [Internet]. 2015 Mar;65(2):87-108. Available from: http://doi.wiley.com/10.3322/caac.21262
- Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology [Internet]. 2013 Jul;58(1):120-7. Available from: http://doi.wiley.com/10.1002/hep.26319
- Jiang J-W, Chen X-H, Ren Z-G, Zheng S-S. Gut microbial dysbiosis associates hepatocellular carcinoma via the gut-liver axis. Hepatobiliary Pancreat Dis Int [Internet]. 2018 Nov; Available from: https://linkinghub.elsevier.com/retrieve/pii/S1499387218302601
- Shen F, Zheng RD, Sun XQ, Ding WJ, Wang XY, Fan JG. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int [Internet]. 2017 Aug;16(4):375-81. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1499387217600195
- Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology [Internet]. 2016 Mar;63(3):764-75. Available from: http://doi.wiley.com/10.1002/hep.28356
- Schwabe RF, Greten TF. Gut microbiome in HCC – Mechanisms, diagnosis and therapy. J Hepatol [Internet]. 2020 Feb;72(2):230-8. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0168827819304830
- Zhou R, Fan X, Schnabl B. Role of the intestinal microbiome in liver fibrosis development and new treatment strategies. Transl Res [Internet]. 2019 Jul;209:22-38. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1931524419300374
- Roderburg C, Luedde T. The role of the gut microbiome in the development and progression of liver cirrhosis and hepatocellular carcinoma. Gut Microbes. 2014.
- Tao X, Wang N, Qin W. Gut Microbiota and Hepatocellular Carcinoma. Gastrointest Tumors. 2015.
- Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, et al. Promotion of Hepatocellular Carcinoma by the Intestinal Microbiota and TLR4. Cancer Cell [Internet]. 2012 Apr;21(4):504-16. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1535610812000724
- Pimentel M. Review of rifaximin as treatment for SIBO and IBS. Expert Opin Investig Drugs [Internet]. 2009 Mar 9;18(3):349-58. Available from: http://www.tandfonline.com/doi/full/10.1517/
13543780902780175 - Shayto RH, Abou Mrad R, Sharara AI. Use of rifaximin in gastrointestinal and liver diseases. World J Gastroenterol [Internet]. 2016;22(29):6638-51. Available from: http://www.wjgnet.com/1007-9327/full/v22/i29/6638.htm
- Abdel-Razik A, Mousa N, Shabana W, Refaey M, Elzehery R, Elhelaly R, et al. Rifaximin in nonalcoholic fatty liver disease: Hit multiple targets with a single shot. Eur J Gastroenterol Hepatol [Internet]. 2018 Oct;30(10):1237-46. Available from: http://journals.lww.com/00042737-201810000-00019
- Cobbold JFL, Atkinson S, Marchesi JR, Smith A, Wai SN, Stove J, et al. Rifaximin in non-alcoholic steatohepatitis: An open-label pilot study. Hepatol Res [Internet]. 2018 Jan;48(1):69-77. Available from: http://doi.wiley.com/10.1111/hepr.12904
- Gangarapu V, Ince AT, Baysal B, Kayar Y, Kiliç U, Gök Ö, et al. Efficacy of rifaximin on circulating endotoxins and cytokines in patients with nonalcoholic fatty liver disease. Eur J Gastroenterol Hepatol [Internet]. 2015 Jul;27(7):840-5. Available from: http://content.wkhealth.com/
linkback/openurl?sid=WKPTLP:landingpage&an=00042737-
201507000-00012 - de Lima VMR, Oliveira CPMS, Alves VAF, Chammas MC, Oliveira EP, Stefano JT, et al. A rodent model of NASH with cirrhosis, oval cell proliferation and hepatocellular
carcinoma. J Hepatol [Internet]. 2008 Dec;49(6):1055-61. Available from: https://linkinghub.elsevier.com/retrieve/pii/S016882780800562X - Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology [Internet]. 2005 Jun;41(6):1313-21. Available from: http://doi.wiley.com/10.1002/hep.20701
- Edmondson HA, Steiner PE. Primary carcinoma of the liver. A study of 100 cases among 48,900 necropsies. Cancer [Internet]. 1954 May;7(3):462-503. Available from: http://doi.wiley.com/10.
1002/1097-0142%28195405%297%3A3%3C462%3A%3AAID-CNCR2820070308%3E3.0.CO%3B2-E - Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol [Internet]. 2011;12(6):R60. Available from: http://genomebiology.biomedcentral.com/articles/10.1186/gb-2011-12-6-r60
- Carvalho CF, Chammas MC, Souza de Oliveira CPM, Cogliati B, Carrilho FJ, Cerri GG. Elastography and Contrast-enhanced Ultrasonography in the Early Detection of Hepatocellular Carcinoma in an Experimental Model of Nonalcoholic Steatohepatitis. J Clin Exp Hepatol. 2013.
- Ezzaidi N, Zhang X, Coker OO, Yu J. New insights and therapeutic implication of gut microbiota in non-alcoholic fatty liver disease and its associated liver cancer. Cancer Lett [Internet]. 2019 Sep;459:186-91. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0304383519303477
- Temraz S, Nassar F, Kreidieh F, Mukherji D, Shamseddine A, Nasr R. Hepatocellular Carcinoma Immunotherapy and the Potential Influence of Gut Microbiome. Int J Mol Sci [Internet]. 2021 Jul 21;22(15):7800. Available from: https://www.mdpi.com/1422-0067/22/15/7800
- Chu H, Williams B, Schnabl B. Gut microbiota, fatty liver disease, and hepatocellular carcinoma. Liver Res [Internet]. 2018 Mar;2(1):43-51. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2542568418000041
- Pinzone MR, Celesia BM, Di Rosa M, Cacopardo B, Nunnari G. Microbial Translocation in Chronic Liver Diseases. Int J Microbiol [Internet]. 2012;2012:1–12. Available from: http://www.hindawi.com/journals/ijmicro/2012/694629/
- Ginès P. Simvastatin Plus Rifaximin in Decompensated Cirrhosis (LIVERHOPE) [Internet]. ClinicalTrials.gov. 2018 [cited 2022 Apr 22]. Available from: https://clinicaltrials.gov/ct2/show/NCT03150459
- Vajro P, Mandato C, Licenziati MR, Franzese A, Vitale DF, Lenta S, et al. Effects of Lactobacillus rhamnosus strain GG in pediatric obesity-related liver disease. J Pediatr Gastroenterol Nutr [Internet]. 2011;52(6):740-3. Available from: https://www.ncbi.nlm.nih.gov/pubmed/21505361
- Holmes E, Li J V., Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab [Internet]. 2012 Nov;16(5):559-64. Available from: https://linkinghub.elsevier.com/retrieve/pii/S155041311200407X
- Wu L, Feng J, Li J, Yu Q, Ji J, Wu J, et al. The gut microbiome-bile acid axis in hepatocarcinogenesis. Biomed Pharmacother [Internet]. 2021 Jan;133:111036. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0753332220312282
- Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature [Internet]. 2013 Jul 26;499(7456):97-101. Available from: http://www.nature.com/articles/nature12347
- Ponziani FR, Bhoori S, Castelli C, Putignani L, Rivoltini L, Del Chierico F, et al. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology [Internet]. 2019 Jan;69(1):107-20. Available from: http://doi.wiley.com/10.1002/hep.30036
- Ferreira DMS, Afonso MB, Rodrigues PM, Simao AL, Pereira DM, Borralho PM, et al. c-Jun N-Terminal Kinase 1/c-Jun Activation of the p53/MicroRNA 34a/Sirtuin 1 Pathway Contributes to Apoptosis Induced by Deoxycholic Acid in Rat Liver. Mol Cell Biol [Internet]. 2014 Mar 15;34(6):1100-20. Available from: http://mcb.asm.org/cgi/doi/10.1128/MCB.00420-13
- Zhao Y, Wu J, Li J V., Zhou NY, Tang H, Wang Y. Gut microbiota composition modifies fecal metabolic profiles in mice. J Proteome Res [Internet]. 2013 Jun 7;12(6):2987-99. Available from: https://pubs.acs.org/doi/10.1021/pr400263n
- Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology [Internet]. 2011 Aug;54(2):562-72. Available from: http://doi.wiley.com/10.1002/hep.24423
- Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014 May;60(5):940-7.
- Wu W, Lv L, Shi D, Ye J, Fang D, Guo F, et al. Protective Effect of Akkermansia muciniphila against Immune-Mediated Liver Injury in a Mouse Model. Front Microbiol [Internet]. 2017 Sep 26;8(SEP). Available from: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01804/full
- Kim S, Lee Y, Kim Y, Seo Y, Lee H, Ha J, et al. Akkermansia muciniphila Prevents Fatty Liver Disease, Decreases Serum Triglycerides, and Maintains Gut Homeostasis. Drake HL, editor. Appl Environ Microbiol. 2020 Jan;86(7).
- Jiang C, Xie C, Li F, Zhang L, Nichols RG, Krausz KW, et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest [Internet]. 2015 Jan 2;125(1):386-402. Available from: http://www.jci.org/articles/view/76738
- Bergheim I, Weber S, Vos M, Krämer S, Volynets V, Kaserouni S, et al. Antibiotics protect against fructose-induced hepatic lipid accumulation in mice: Role of endotoxin. J Hepatol [Internet]. 2008 Jun;48(6):983-92. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0168827808001323
- Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature [Internet]. 2012 Feb 1;482(7384):179–85. Available from: http://www.nature.com/articles/nature10809
- Yamada S, Kamada N, Amiya T, Nakamoto N, Nakaoka T, Kimura M, et al. Gut microbiota-mediated generation of saturated fatty acids elicits inflammation in the liver in murine high-fat diet-induced steatohepatitis. BMC Gastroenterol [Internet]. 2017 Dec 29;17(1):136. Available from: https://bmcgastroenterol.biomedcentral.com/articles/10.1186/s12876-017-0689-3
- Lv XY, Ding HG, Zheng JF, Fan CL, Li L. Rifaximin improves survival in cirrhotic patients with refractory ascites: A real-world study. World J Gastroenterol. 2020;26(8):199-218.
- Fodor AA, Pimentel M, Chey WD, Lembo A, Golden PL, Israel RJ, et al. Rifaximin is associated with modest, transient decreases in multiple taxa in the gut microbiota of patients with diarrhoea-predominant irritable bowel syndrome. Gut Microbes [Internet]. 2019 Jan 2;10(1):22-33. Available from: https://www.tandfonline.com/doi/full/10.1080/19490976.2018.1460013
- Ponziani FR, Zocco MA, D’Aversa F, Pompili M, Gasbarrini A. Eubiotic properties of rifaximin: Disruption of the traditional concepts in gut microbiota modulation. World J Gastroenterol [Internet]. 2017;23(25):4491. Available from: http://www.wjgnet.com/1007-9327/full/v23/i25/4491.htm
- Bajaj JS, Sikaroodi M, Shamsaddini A, Henseler Z, Santiago-Rodriguez T, Acharya C, et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut [Internet]. 2021 Jun;70(6):1162-73. Available from: https://gut.bmj.com/lookup/doi/10.1136/gutjnl-2020-322470
- Jiang L, Chu H, Gao B, Lang S, Wang Y, Duan Y, et al. Transcriptomic Profiling Identifies Novel Hepatic and Intestinal Genes Following Chronic Plus Binge Ethanol Feeding in Mice. Dig Dis Sci [Internet]. 2020 Dec 15;65(12):3592-604. Available from: https://link.springer.com/10.1007/s10620-020-06461-6
- Li F, Ye J, Shao C, Zhong B. Compositional alterations of gut microbiota in nonalcoholic fatty liver disease patients: a systematic review and Meta-analysis. Lipids Health Dis [Internet]. 2021 Dec 26;20(1):22. Available from: https://lipidworld.biomedcentral.com/articles/10.1186/s12944-021-01440-w
Correspondence: Carolina Uribe-Cruz
Email: carolinaurib10@yahoo.com.ar
Acta Gastroenterol Latinoam 2023;53(3):265-282
Supplementary data
Materials and Methods
Probes Identification
Supplementary Table 1. Describes the identification of TaqMan probes used to evaluate the gene expression

Bioinformatics analyses
Processing of 16S rRNA reads for downstream analyses
Sequence data exported from the Ion PGM™ system were processed using a custom pipeline in Mothur v.1.41.1.1 First, sequences were barcodes and primers depleted (no mismatch allowed) and then a quality filter was applied to eliminate low quality reads. Quality control was performed by trimming low quality reads (those of incorrect length, containing an ambiguous base or containing homopolymers longer than 8 bp). All potentially chimeric sequences were identified and removed using VSEARCH.2
After these initial quality filtering and trimming steps, the remaining sequences were clustered into operational taxonomic units (OTUs) at a 99% identity level and classified using the SILVA v132 reference database at 97% similarity.3 Sequences that could not be classified (i.e., “unknown” sequences), as well as sequences identified as eukaryotes, mitochondria and chloroplasts, were removed prior to further analysis. To reduce spurious OTUs caused by PCR or sequencing errors, an additional filtering step was performed by removing OTUs with fewer than 10 reads. The resulting OTU table was normalized using the cumulative sum scaling (CSS) method. For alpha diversity analysis, the OTU table was rarefied to the smallest library size. Subsequent analyses of the sequence dataset were performed in R v. 3.6.1 (using the vegan, phyloseq, ggplot 2 and Microbiome Analyst R packages).
Results
Supplementary Table 2. Body weight
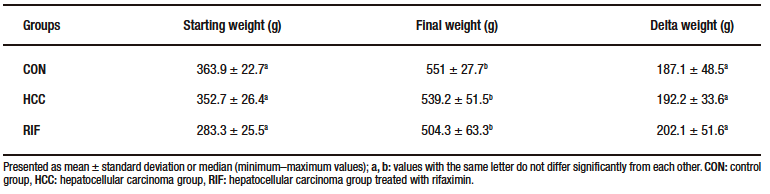
Supplementary Table 3. Biochemical parameters
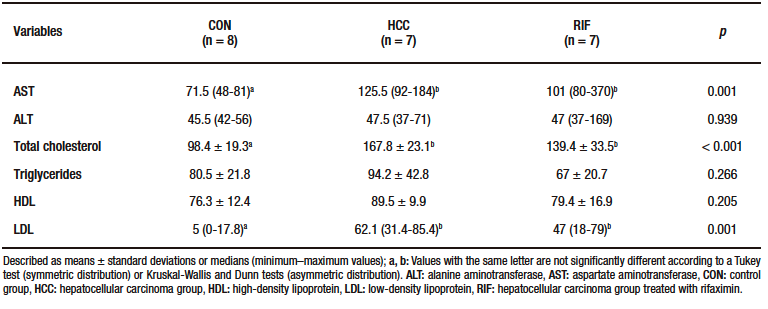
Supplementary Figure 1. Hepatic gene expression of pro- and anti-inflammatory cytokines
Supplementary Figure 2. Gene expression of intestinal permeability markers
Supplementary Figure 3. Assessment of liver fibrosis
Supplementary Figure 4. Composition of the gut microbiota in phyla, families and genera
Supplementary Table 4. Distribution of histologic findings of fibrosis scores
Supplementary Table 5. Distribution of histological findings of cancer grading
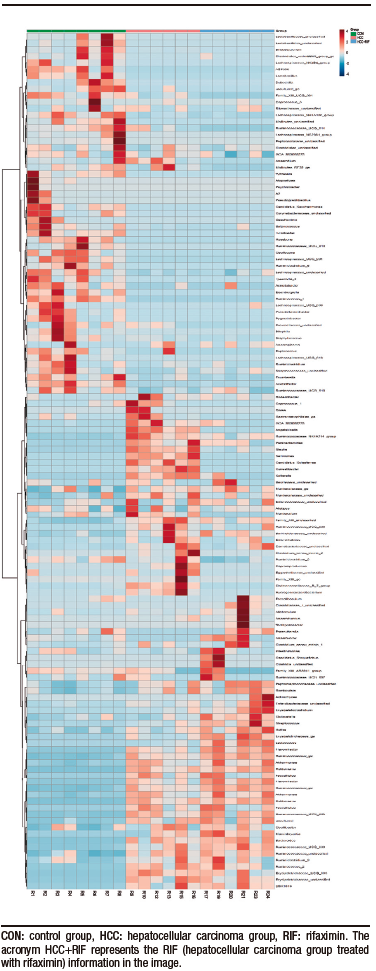
Supplementary Figure 5. Distribution of the 124 identified taxonomic genera among the samples
References
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol [Internet]. 2009 Dec 1;75(23):7537-41. Available from: https://aem.asm.org/content/75/23/7537
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: A versatile open source tool for metagenomics. PeerJ [Internet]. 2016 Oct 18;2016(10):e2584. Available from: https://peerj.com/articles/2584
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res [Internet]. 2013 Nov 27;41(D1):D590–6. Available from: http://academic.oup.com/nar/article/41/D1/D590/1069277/The-SILVA-ribosomal-RNA-gene-database-project
 Revista ACTA Órgano Oficial de SAGE
Revista ACTA Órgano Oficial de SAGE